Photometry
To measure the Petrosian properties of galaxies, we construct a photometric curve of growth using a set of concentric apertures with varying radii to measure the flux. From this curve of growth, we will measure different properties of the galaxy including the total flux, characteristic radii such as the Petrosian radius and half-light radius, and the concentration index in the Petrosian section below. For a quick guide on how to construct curves of growth and Petrosian profiles, please see the Making a Photutils Source Catalog and Curve of Growth and Petrosian Radii sections in the Quick Start guide.
To start with PetroFit, simply import it as follows:
[1]:
import petrofit as pf
Loading Example Data
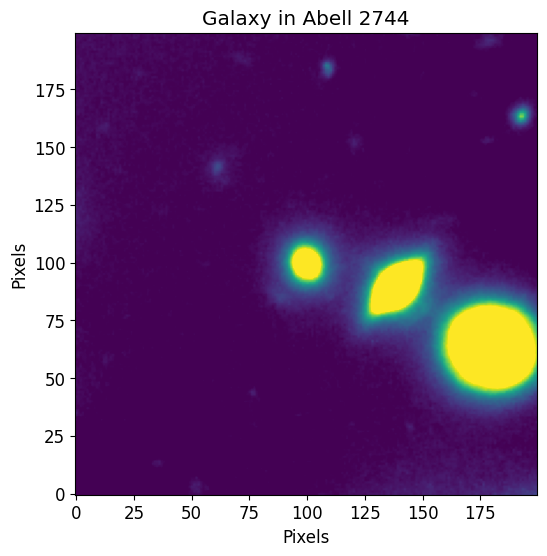
The following data is a cutout of a group of bright galaxies in Abell 2744 (located at (3.596248, -30.388517)). The original data was acquired by the Hubble Frontier Fields team via the WFC3 instrument in the F105W filter and can be directly downloaded from the Mikulski Archive for Space Telescopes. The cutout image used
in this documentation can be found in the git repository at the following path petrofit/docs/data/abell_2744_dwarf_galaxy_f105w.fits.gz.
We first use astropy’s CCDData to load the example data and visualize it through matplotlib.
[2]:
from astropy.nddata import CCDData
from astropy.io import fits
image = CCDData.read('data/abell_2744_dwarf_galaxy_f105w.fits.gz')
rms = fits.getdata('data/abell_2744_dwarf_galaxy_f105w_rms.fits.gz')
[4]:
from matplotlib import pyplot as plt
plt.rcParams['figure.figsize'] = [6, 6]
plt.rcParams['image.origin'] = 'lower'
plt.rcParams['font.size'] = 12
vmax = image.data.std() # Use the image std as max and min of all plots
vmin = - vmax
plt.imshow(image.data, vmin=0, vmax=image.data.std())
plt.title("Galaxy in Abell 2744")
plt.xlabel("Pixels")
plt.ylabel("Pixels")
plt.show()
Estimate Data Noise at Dark Area
In this section, we estimate the noise levels in the image using astropy’s sigma_clipped_stats and photutils’s calc_total_error functions.
[5]:
from astropy.stats import sigma_clipped_stats
image_mean, image_median, image_stddev = sigma_clipped_stats(image.data, sigma=3)
[6]:
from photutils.utils import calc_total_error
err = calc_total_error(
data=image.data, # Input Image
bkg_error=rms, # All sources of background error except source Poisson error
effective_gain=image.header['EXPTIME'] # Factor to convert data units to counts
)
Catalogs
Before we can perform photometry, we need to first identify the sources in the image. To do this, we use Photutils and the wrappers in PetroFit.
Make Catalog
We first start by defining the detection threshold and we select this value to be 3-sigma for this example.
[7]:
# Define detect threshold
threshold = 3*image_stddev
We also have to define the number of pixels that make up the smallest object. npixels is the number of connected pixels, each greater than the threshold value, that an object must have to be detected. npixels must be a positive integer.
[8]:
npixels = 4**2
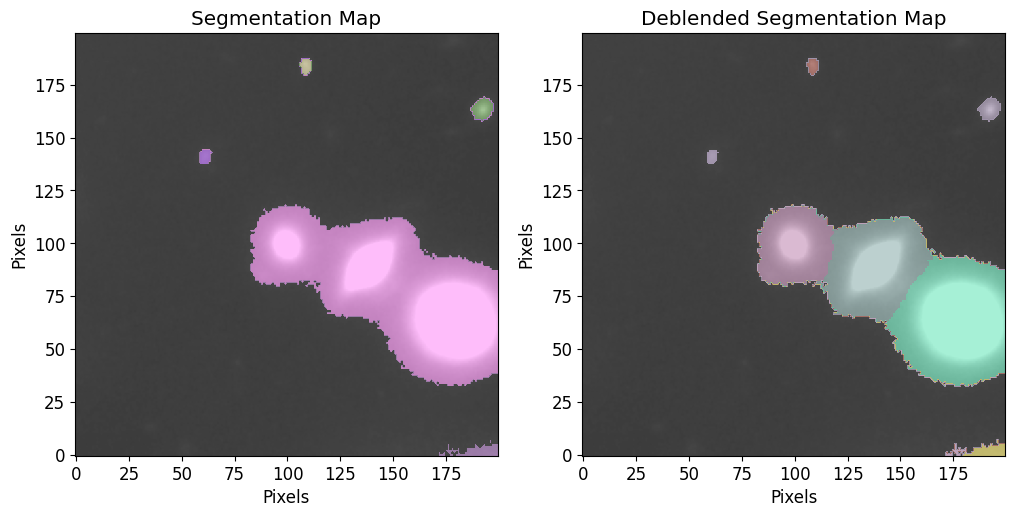
To make a catalog of sources and segmentation maps, we use petrofit’s make_catalog function. The function returns a source catalog (photutils.SourceCatalog), a segmentation image, and, if deblend=True, a deblended segmentation image.
The make_catalog function wraps three steps into one function:
Segmentation:
To identify sources in the image, we first segment the image. The image is smoothed with a gaussian kernel if
kernel_sizeis provided and clipped at the threshold specified (after smoothing if applicable). The image is then segmented using thepetrofit.segmentation.make_segmentsfunction, which is a wrapper forphotutil’sdetect_sourcesfunctionality.Deblending:
To further distinguish the sources, we use
photutils.deblend_sourcesto deblend the sources into individual galaxies. Thecontrastparameter is the fraction of the total (blended) source flux that a local peak must have (at any one of the multi-thresholds) to be considered as a separate object andnlevelsis the number of multi-thresholding levels to use. Ifdeblendis set toFalse,Noneis returend forsegm_deblend.Source Catalog:
Now that we have deblended the sourecs into individual sources, the next step is to create a source catalog (
photutils.SourceCatalog) that contains properties like(xcentroid, ycentroid),eccentricityandarea. Note that the deblended map is used to make the source catalog but ifdeblendis set toFalse, the segmentation map is used instead.
By setting the plot flag to True, we see plots of the segmentation and deblended segmentation maps. The image is plotted along with a color-coded overplot of the segmentation map (each source is a different color). vmax and vmin can be used the same way as in plt.imshow.
After the computation, the following objects are returned:
cat: A catalog of sources.segm: Segmentation map.segm_deblend: Deblended segmentation map.
[9]:
cat, segm, segm_deblend = pf.make_catalog(
image.data,
threshold,
wcs=None,
deblend=True,
npixels=npixels,
nlevels=30,
contrast=0.001,
plot=True, vmax=vmax, vmin=vmin,
figsize=(12, 6)
)
To demonstrate the useful information in the catalog, we convert the SourceCatalog to an astropy.table.Table and display the first 10 objects.
[10]:
# Display source properties
print("Num of Targets:", len(cat))
# Convert to table
cat_table = cat.to_table()
cat_table[:10]
Num of Targets: 8
[10]:
| label | xcentroid | ycentroid | sky_centroid | bbox_xmin | bbox_xmax | bbox_ymin | bbox_ymax | area | semimajor_sigma | semiminor_sigma | orientation | eccentricity | min_value | max_value | local_background | segment_flux | segment_fluxerr | kron_flux | kron_fluxerr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pix2 | pix | pix | deg | ||||||||||||||||
| int64 | float64 | float64 | object | int64 | int64 | int64 | int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
| 1 | 61.08347174238899 | 141.05186559012205 | None | 59 | 64 | 138 | 144 | 34.0 | 1.7994370638925237 | 1.2826901873829257 | 72.98283673407356 | 0.7013382917172669 | 0.010554540902376175 | 0.026845833286643028 | 0.0 | 0.5834723142907023 | nan | 1.8317878434947907 | nan |
| 2 | 192.36299560404697 | 163.40391871827393 | None | 187 | 197 | 158 | 169 | 86.0 | 2.3144983589944554 | 1.9834471217713212 | 53.65520486998864 | 0.5153724293039628 | 0.010591089725494385 | 0.0929785966873169 | 0.0 | 2.8758262349292636 | nan | 3.2707537413335355 | nan |
| 3 | 108.72693550508657 | 183.8851479597333 | None | 106 | 111 | 180 | 187 | 37.0 | 1.8003927404449662 | 1.257057560548163 | -87.14736406479223 | 0.7158901102937193 | 0.010464809834957123 | 0.045437149703502655 | 0.0 | 0.8372195269912481 | nan | 1.0690440471675258 | nan |
| 4 | 176.59884904430535 | 1.7105077906435575 | None | 172 | 181 | 0 | 5 | 22.0 | 2.5652493504083584 | 1.2345618770639404 | 13.128240218386493 | 0.876575763829376 | 0.010684505105018616 | 0.01434874627739191 | 0.0 | 0.26085690688341856 | nan | 3.44449120866383 | nan |
| 5 | 192.09296727764394 | 1.82553577948899 | None | 180 | 199 | 0 | 7 | 84.0 | 4.976606287872485 | 1.5422939287464803 | 5.980651162675711 | 0.9507662987626069 | 0.010602000169456005 | 0.02371048368513584 | 0.0 | 1.247399945743382 | nan | 3.590573711610179 | nan |
| 6 | 178.73633119059068 | 63.493826518219336 | None | 143 | 199 | 33 | 95 | 2704.0 | 9.40299866437019 | 9.177157386885888 | -48.66212288913661 | 0.21785120428951427 | 0.010437765158712864 | 3.6771397590637207 | 0.0 | 421.7741427142173 | nan | 422.6815556552462 | nan |
| 7 | 138.55898473281684 | 89.25004484979968 | None | 109 | 166 | 63 | 112 | 1864.0 | 7.532153967377171 | 5.563519557842017 | 44.31309259442659 | 0.6741045627289916 | 0.010432953014969826 | 3.6545557975769043 | 0.0 | 244.33296140562743 | nan | 240.57513423073874 | nan |
| 8 | 99.92859014542458 | 99.15188824983441 | None | 83 | 118 | 80 | 118 | 1114.0 | 6.7507255757709 | 6.379388610260266 | 76.6082699457646 | 0.32709046915385503 | 0.010427203960716724 | 0.44580602645874023 | 0.0 | 56.775723142549396 | nan | 60.32600669722427 | nan |
Plotting Segmentation Maps
To plot segmentations, you can use the plot_segments function included in PetroFit as follows:
[11]:
pf.plot_segments(segm, image=image.data, vmax=vmax, vmin=vmin)
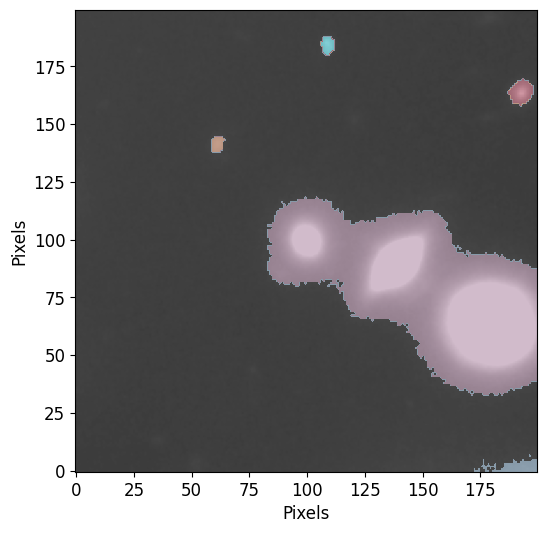
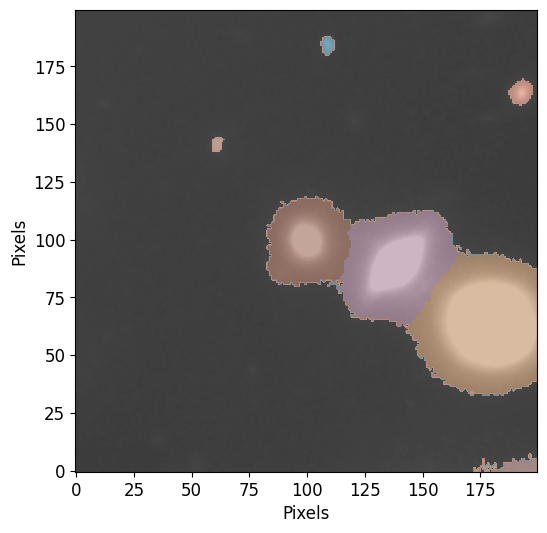
As you can see, the segmentation resulted in the identification sources but the sources at the center were classified as a single object because they have interconnecting pixels that are above the threshold.
Next we use the same functiton to plot the deblended segmentation map, notice how the central sources are now deblended into individual sources:
[12]:
pf.plot_segments(segm_deblend, image=image.data, vmax=vmax, vmin=vmin)
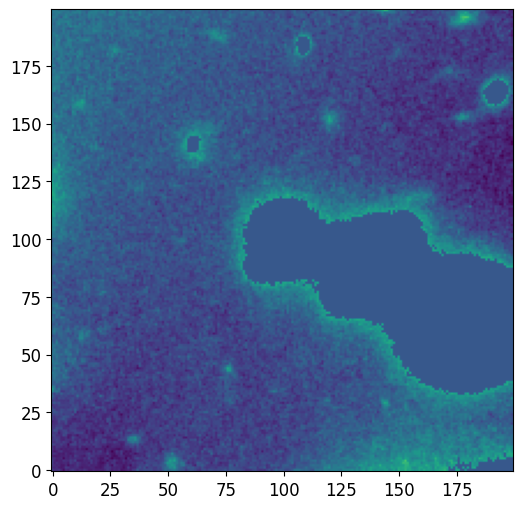
We can also plot the background pixels that are not a part of a source’s segmentation footprint using the plot_segment_residual function. We significantly lower the vmax and vmin values so the background pixels become more apparent. This plot can be used to see if the threshold used to segment the image was too high. If the threshold is high, we would notice bright pixels that are part of the source in this plot.
[13]:
pf.plot_segment_residual(segm, image.data, vmax=vmax/5)
Photometry on Single Source
The purpose of this step is to perform aperture photometry to construct a curve of growth that we can use for the Petrosian measurements.
Source Selection
For this example, we will focus on a single source. We have included a helper function order_cat that will produce a list of indices sorted by a key (default is ‘area’). We use the order_cat function to identify the source of interest and perform photometry on its cutout.
[14]:
# Sort and get the largest object in the catalog
sorted_idx_list = pf.order_cat(cat, key='area', reverse=True)
idx = sorted_idx_list[1] # index 0 is largest
source = cat[idx] # get source from the catalog
Aperture Radii
To construct the curve of growth, we measure the photometry using circular and/or elliptical apertures of varying, concentric radii that are centered on the source. Before we perform the photometry, we need to provide a list of radii that will be used to construct the circular and elliptical apertures. To achieve this, we have provided a helper function that takes in the max radius in pixels (max_pix) and the number radii (n). The function will return a list of radii by dividing the
range (max_pix/n, max_pix) into n equally spaced integers.
[15]:
r_list = pf.make_radius_list(
max_pix=50, # Max pixel to go up to
n=50 # the number of radii to produce
)
print(repr(r_list))
array([ 1., 2., 3., 4., 5., 6., 7., 8., 9., 10., 11., 12., 13.,
14., 15., 16., 17., 18., 19., 20., 21., 22., 23., 24., 25., 26.,
27., 28., 29., 30., 31., 32., 33., 34., 35., 36., 37., 38., 39.,
40., 41., 42., 43., 44., 45., 46., 47., 48., 49., 50.])
Photometry Calculation
The source_photometry function is used to perform photometry on a single source (photutils.segmentation.catalog.SourceCatalog). In this example, we provide the function with the source object, the raw image (note that this is the 2D array and not CCDData), segmentation from the last step (SegmentationImage) and the list of radii we made using make_radius_list. Given these parameters and the options below the function performs photometry as follows:
The positions (max value of source in pixels), elongation and position-angle of the source are determined from the
SourceCatalogobject. The position will be used to center the apertures and the elongation and position angles will be used as parameters of the elliptical apertures. Each of the radii will be assigned an elliptical aperture with these parameters.If
cutout_sizeis defined, the code will use it to make a cutout of that size with the source centered. If thecutout_sizeis larger than the image or contains pixels outside the image, those pixels outside of the image are replaced bynp.nan.Once the
cutout_sizeis determined, cutouts of the error map (if provided) and image are produced. Before the raw image is cutout, sources that are not the source of interest are masked using the segmentation map. Themask_backgroundoption gives us the ability to also mask pixels that are considered to be background pixels because they do not belong to any source’s segmentation map. All masked pixels are replaced bynp.nanand are not counted in the returned area array.If
bg_subis set to true, a 2D plane is used to fit pixels that are below a specified sigma from the mean using thepetrofit.fitting.fit_planefunction. The sigmasigmavalue is used to determine noise pixels. Once the pixels above this value are masked, a 2D plane is fit to determine the background. The 2D plane model is then converted into an image and subtracted from the cutout of the target source.sigma_typeis used to set how thissigmavalue will be used. Thesigma_typeoptions are'clip'and'bound':'clip'(default): Usesastropy.stats.sigma_clipping.sigma_clipto clip at the providedsigmastd value. Note thatsigmain this case is the number of stds above the mean.'bound': After computing the mean of the image, clip atmean - sigmaandmean + sigma. Note thatsigmain this case is a value and not the number of stds above the mean.
The resulting image (after being noise subtracted if
bg_subis set to true) is passed to thepetrofit.photometry.photometry_stepwhich constructs the apertures and performs photometry.
After calculating the photometry at each radius, three arrays are returned:
flux_arr: Photometric sum in aperture.area_arr: Exact area of the aperture.error_arr: if error map is provided, error of measurements.
[16]:
# Photomerty
flux_arr, area_arr, error_arr = pf.source_photometry(
# Inputs
source, # Source (`photutils.segmentation.catalog.SourceCatalog`)
image.data, # Image as 2D array
segm_deblend, # Deblended segmentation map of image
r_list, # list of aperture radii
error=err,
# Options
cutout_size=max(r_list)*2, # Cutout out size, set to double the max radius
bg_sub=True, # Subtract background
sigma=3, sigma_type='clip', # Fit a 2D plane to pixels within 3 sigma of the mean
plot=True, vmax=vmax, vmin=vmin, # Show plot with max and min defined above
)
plt.show()
7
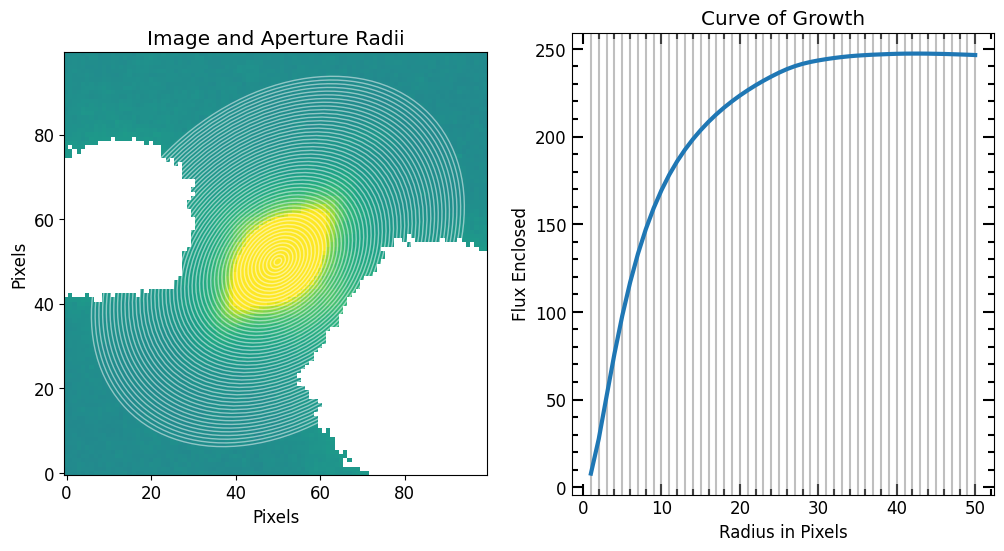
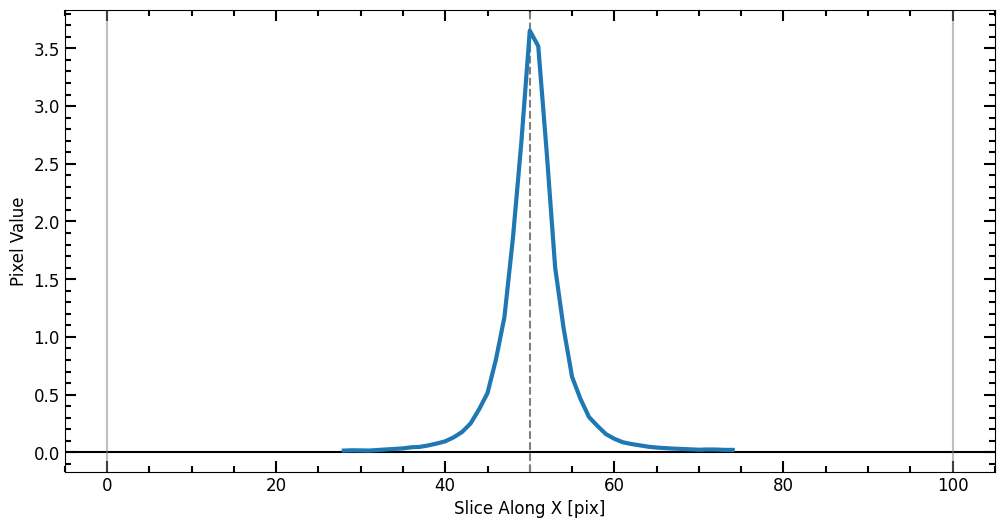
If source_photometry’s plot option is set to True, four plots are displayed: - The top left plot shows the cutout with the 2D plane background subtraction and the surrounding sources masked (replaced by np.nan). - The top right plot shows the curve of growth with increasing radius. The red lines represent the aperture radius. - The bottom two plots show the source profile sliced at the center of the image in the y and x direction respectively.
Save Photometry Arrays
There are many ways of saving the photometry results. For example adding the photometry to the source catalog with the radii as columns. One simple way to save the results for a single source is to save it as a csv of ecsv file using AstroPy Table.
[17]:
from astropy.table import Table
t = Table(
data=[r_list, flux_arr, area_arr, error_arr],
names=['r_list', 'flux_arr', 'area_arr', 'error_arr'],
)
t[:10] # Show first 10 radii
[17]:
| r_list | flux_arr | area_arr | error_arr |
|---|---|---|---|
| float64 | float64 | float64 | float64 |
| 1.0 | 7.753508 | 2.320493 | 0.011168 |
| 2.0 | 27.470488 | 9.281973 | 0.021141 |
| 3.0 | 51.868013 | 20.884439 | 0.029355 |
| 4.0 | 76.013873 | 37.127891 | 0.03601 |
| 5.0 | 97.787811 | 58.01233 | 0.041509 |
| 6.0 | 116.781632 | 83.537756 | 0.046185 |
| 7.0 | 133.120986 | 113.704168 | 0.050296 |
| 8.0 | 147.100726 | 148.511566 | 0.054002 |
| 9.0 | 159.118392 | 187.95995 | 0.057433 |
| 10.0 | 169.405595 | 232.049321 | 0.060678 |
[18]:
# also save shape information in the table's meta
t.meta['position'] = pf.get_source_position(source)
t.meta['elong'] = pf.get_source_elong(source)
t.meta['theta'] = pf.get_source_theta(source)
[19]:
t.write('temp/abell_2744_galaxy_f105w_photometry.ecsv', overwrite=True)
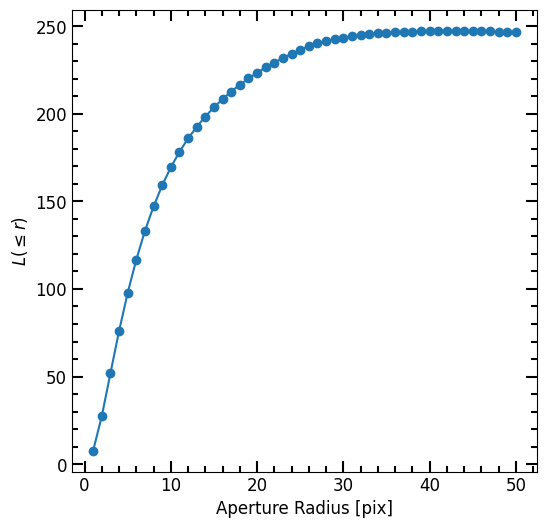
Plot Curve of Growth
Here we plot the curve of growth:
[20]:
plt.errorbar(r_list, flux_arr, yerr=error_arr,
marker='o', capsize=3, label="Data")
pf.mpl_tick_frame()
plt.xlabel("Aperture Radius [pix]")
plt.ylabel("$L(\leq r)$")
plt.show()