Correction Grids
Correction grids can be thought of as lookup tables that provide corrected Petrosian derived values. These grids are typically generated through simulations or theoretical models to offer the best correction for Petrosian profiles. This section discusses the concept of correction grids: how they’re generated, when and why they should be used, and how they enhance accuracy.
Necessity and Implications
Are Correction Grids Always Needed?
The short answer is no, but they are very useful in some cases. Petrosian corrections offer more precise measurements of galaxies. The necessity hinges on your data quality, research goals, and desired precision. Here are key considerations on why the defualt settings are usually good enough:
Standard Practices: Numerous studies, such as those from the Sloan Digital Sky Survey (SDSS), rely on default settings. The parameters \(\eta = 0.2\) and \(\epsilon = 2\) are widely accepted because they typically produce reliable results across many scenarios.
Noise Constraints: If the fainter parts of galaxies in your images are submerged below the noise level, detailed corrections might not significantly impact the outcome since that portion of the galaxy is not meaningfully measurable. Here, the additional steps and computational demands of correction grids might be unwarranted.
Practical Implications: While correction grids can produce improved results, they require optimal background subtraction. Not all datasets might meet this standard.
Correction grids promise enhanced precision but aren’t always essential. For a majority of cases, default settings suffice.
When are Correction Grids Needed?
While the default settings serve most general purposes, there are specific scenarios where correction grids are beneficial:
Galaxy Size Estimation: If your research aims to measure the size of a galaxy which suffers due to poor signal-to-noise, you can estimate its total flux radius based on the corrected epsilon. Remember, this isn’t about recovering the total flux which includes regions submerged under noise, but about the estimating the full extent of the galaxy based on the measurable components of the light profile.
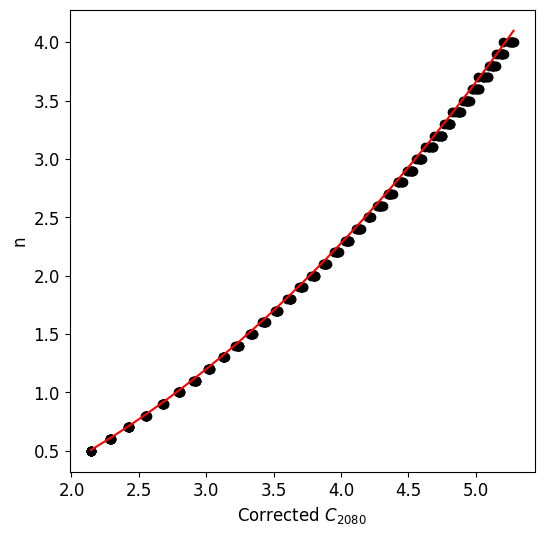
Estimating Sérsic Index: Correction grids can be used to estimate the Sérsic index of single component galaxies.
High-Precision Studies: For projects that require a granular study of galaxy structures, or when analyzing galaxies with distinct features that might get overlooked with standard parameters, correction grids provide the extra precision that may be needed.
Superior Data Quality: If you have high-quality images with excellent background subtraction and reasonable signal-to-noise ratios, the total flux can be estimated to higher accuracy. That is, high quality data allows you to measure the flux up to the corrected total flux radius.
In essence, when the goal shifts from general measurements to intricate, high-resolution studies of galactic structures and sizes, correction grids become a useful tool.
Generating a Correction Grid
Correction grids are essentially lookup tables that map certain observational properties of a galaxy (like Petrosian radius, uncorrected half light radius, and concentration indices) to more intrinsic properties (like Sérsic index and the corrected epsilon value). In this section, we’ll walk through the steps of creating a correction grid.
To start with PetroFit, simply import it as follows:
[2]:
import petrofit as pf
Load PSF (Optional)
We load an HST F105W PSF and normalize it to sum to 1:
[3]:
from matplotlib import pyplot as plt
plt.rcParams['figure.figsize'] = [6, 6]
plt.rcParams['image.origin'] = 'lower'
plt.rcParams['font.size'] = 12
[4]:
from astropy.io import fits
# Load PSF image (2D array)
PSF = fits.getdata('data/f105w_psf.fits.gz')
# Normalize PSF
PSF = PSF / PSF.sum()
# Note that the PSF shape is odd on all sides
print("PSF Shape = {}".format(PSF.shape))
# Plot PSF and use vmax and vmin to show difraction spikes
plt.imshow(PSF, vmin=0, vmax=5e-4)
plt.show()
PSF Shape = (50, 50)
Define Sampling Points
The correction grid is generated along with a set of half-light radii and Sersic indices. The generator loops through the Sersic indices for each half-light radius in the radius list. For this documentation, we define a small set of values, with the Sersic indices of a Gaussian and de Vaucouleurs’ profile.
[5]:
import numpy as np
r_eff_list = np.array([7, 10, 15])
n_list = np.array([0.5, 1., 4.])
Grid Simulation
We do a simple API call to generate the correction grid. We provide a PSF and oversampling rule as well. Oversampling becomes important for small half-light radii since the model needs to be sampled well in the center.
The generate_petrosian_sersic_correction grid follows the following steps to generate the correction grid:
Computes the total flux (
total_flux) of an ideal Sersic profile with the sampling points using thepetrofit.modeling.models.sersic_enclosedfunction and setting the radius tonp.inf.Computes the radius equal to
total_flux * 0.99.Makes a PSF convolved Sersic Model image and measures the photometry.
Measures the uncorrected Petrosian radius.
Computes the correct
epsilonvalue ascorrected_epsilon = r(total_flux) / r_petrosian.It also saves other values, such as the uncorrected
C2080which can be used to mapto the Sersic index.
[6]:
output_file_name='temp/example_correction_gid.ecsv'
petrosian_grid = pf.generate_petrosian_sersic_correction(
output_file_name=output_file_name, # Output file name
psf=PSF, # PSF (optional)
r_eff_list=r_eff_list, # List of r_e to sample
n_list=n_list, # List of n to sample
oversample=4, # Oversample factor or region, see fitting docs
out_format='ascii.ecsv', # Output format, should match file name
overwrite=True, # Overwrite output
ipython_widget=True, # Progress bar
n_cpu=None, # int value for number of mp threads
plot=False, # Plot each step, can not be True if n_cpu > 1
)
[7]:
petrosian_grid
[7]:
| n | r_eff | sersic_r_20 | sersic_r_80 | sersic_r_99 | sersic_L_inf | p02 | p03 | p04 | p05 | p0502 | p0302 | u_epsilon | u_epsilon_50 | u_epsilon_80 | u_r_50 | u_r_99 | u_r_20 | u_r_80 | u_c2080 | u_c5090 | c_epsilon | c_epsilon_50 | c_epsilon_80 | c_r_50 | c_r_99 | c_r_20 | c_r_80 | c_c2080 | c_c5090 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| float64 | int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
| 0.5 | 7 | 3.9494261674515654 | 10.535743151744386 | 18.04297517869382 | 22208.564694953242 | 16.690338067613524 | 14.742948589717944 | 13.191638327665533 | 11.816363272654531 | 0.7499064971159685 | 0.2694039121096147 | 2.0 | 0.4702462995145921 | 0.7427218793072451 | 7.848569713942788 | 33.38487697539508 | 4.402680536107221 | 12.39627925585117 | 2.247870887683088 | 1.477215403931874 | 1.5838256474738557 | 0.4676574579013603 | 0.7294770646895044 | 7.805361072214443 | 26.43908781756351 | 4.3810762152430485 | 12.255851170234047 | 2.2338133500139796 | 1.443311364049927 |
| 0.5 | 10 | 5.6420373820736645 | 15.051061645349122 | 25.77567882670543 | 45323.60141827192 | 22.48849769953991 | 19.885377075415082 | 17.75975195039008 | 15.834366873374675 | 0.761798705625901 | 0.267133067590084 | 2.0 | 0.4798345490126312 | 0.7461928482476425 | 10.790758151630326 | 44.98239647929586 | 6.0778155631126225 | 16.780756151230243 | 2.2053200481575006 | 1.398916647555213 | 1.3667428966087338 | 0.47645436755025794 | 0.729443744213396 | 10.714742948589718 | 30.737147429485894 | 6.047409481896379 | 16.567913582716542 | 2.1884922205394455 | 1.3671755462140593 |
| 0.5 | 15 | 8.463056073110499 | 22.576592468023684 | 38.66351824005819 | 101978.10319111182 | 32.02240448089618 | 28.364672934586917 | 25.30506101220244 | 22.521504300860173 | 0.7642827173570272 | 0.26338079339405823 | 2.0 | 0.4898488255872064 | 0.7547663668165917 | 15.686137227445489 | 64.05461092218442 | 8.86757351470294 | 24.169433886777355 | 2.1773084029854246 | 1.3534094001636479 | 1.2911297807635969 | 0.4862881059470265 | 0.736866400939908 | 15.572114422884576 | 41.341268253650725 | 8.821964392878575 | 23.8501700340068 | 2.1596308951669485 | 1.3247894047084208 |
| 1.0 | 7 | 3.4152455989729886 | 12.269877985786641 | 27.687042520030552 | 10929.805669580057 | 18.977595519103822 | 15.490898179635927 | 12.917383476695338 | 10.841968393678735 | 1.2156652456010393 | 0.4408229332439093 | 2.0 | 0.42609282273450755 | 0.7614605403240257 | 8.08621724344869 | 37.95659131826365 | 4.067413482696539 | 14.450690138027605 | 2.752851323921932 | 1.854400745123529 | 1.6516698747257175 | 0.4226354235841001 | 0.7417938655072812 | 8.020604120824164 | 31.346069213842767 | 4.051010202040407 | 14.25385077015403 | 2.7318443537737207 | 1.8246976026729558 |
| 1.0 | 10 | 4.8789222842471265 | 17.52839712255235 | 39.552917885758006 | 22305.725856285833 | 25.280656131226245 | 20.63872774554911 | 17.068013602720544 | 14.11622324464893 | 1.265349116905938 | 0.44052709634619963 | 2.0 | 0.43544762537783477 | 0.7715820791593473 | 11.008401680336068 | 50.56991398279656 | 5.508501700340068 | 19.506101220244048 | 2.745684928420304 | 1.8121310726092839 | 1.641241793161436 | 0.43264650492965545 | 0.7487453452425344 | 10.9375875175035 | 41.482096419283856 | 5.484896979395879 | 19.24644928985797 | 2.7259107247705625 | 1.7791529983794634 |
| 1.0 | 15 | 7.31838342637069 | 26.292595683828527 | 59.32937682863701 | 50187.88317664311 | 35.513302660532105 | 28.92758551710342 | 23.79355871174235 | 19.473894778955792 | 1.3046912176308711 | 0.44539438244001306 | 2.0 | 0.4465191994637556 | 0.7895353487559919 | 15.857371474294858 | 71.0268053610722 | 7.900580116023204 | 28.03900780156031 | 2.7505183002812106 | 1.8075396077736015 | 1.700997857031393 | 0.4435450710016842 | 0.7676036327672117 | 15.751750350070013 | 60.39427885577115 | 7.865373074614922 | 27.722144428885773 | 2.7355374733767586 | 1.7796966409157084 |
| 4.0 | 7 | 1.9338894613170134 | 21.37899838138344 | 132.60580887212956 | 51.861320264524814 | 20.729545909181834 | 12.735347069413882 | 8.928585717143427 | 6.782956591318263 | 2.4258537551641766 | 1.0578950185789684 | 2.0 | 0.368581547280149 | 0.8897584606328467 | 7.640528105621124 | 41.4624924984997 | 2.926785357071414 | 18.444288857771554 | 3.997355207847013 | 2.6804143543491543 | 7.164423510366633 | 0.42169511806768517 | 1.1173986772316422 | 8.741548309661933 | 148.50230046009202 | 3.167633526705341 | 23.91498299659932 | 4.3896757273607 | 3.312617849643298 |
| 4.0 | 10 | 2.7626992304528764 | 30.541426259119202 | 189.43686981732796 | 105.83942911127511 | 25.805161032206442 | 15.783156631326266 | 10.442088417683536 | 7.621524304860972 | 2.6483236731181488 | 1.0675635354491433 | 2.0 | 0.3794728682170542 | 0.9125736434108527 | 9.792358471694339 | 51.60092018403681 | 3.631726345269054 | 23.549109821964393 | 4.05930689273775 | 2.6732303489915554 | 7.8467121681302565 | 0.4490077519379845 | 1.2233150416937433 | 11.586717343468694 | 202.50650130026006 | 3.990598119623925 | 32.7003400680136 | 4.567571377813081 | 3.4406515906900377 |
| 4.0 | 15 | 4.144048845679315 | 45.8121393886788 | 284.15530472598925 | 238.138715500369 | 33.366673334666935 | 20.404080816163233 | 12.882576515303061 | 8.881776355271054 | 2.8740653809355003 | 1.0679793726990117 | 2.0 | 0.39595923261390886 | 0.9509232613908873 | 13.211842368473695 | 66.76835367073416 | 4.671534306861373 | 31.729145829165837 | 4.159994190872208 | 2.664062476224748 | 8.860372029430788 | 0.48925059952038374 | 1.3916191368348632 | 16.3246649329866 | 295.6805361072215 | 5.310062012402481 | 48.17123424684937 | 4.788440905285227 | 3.559459521262453 |
PetrosianCorrection
The PetrosianCorrection class is designed to compute corrections for Petrosian based on default Petrosian measurements.
Correcting a Petrosian Profile
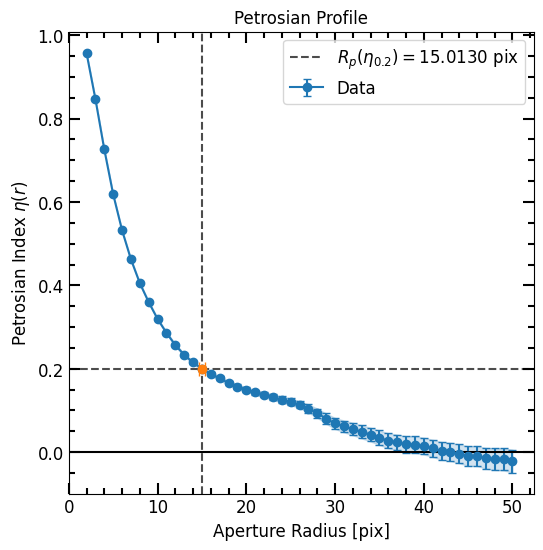
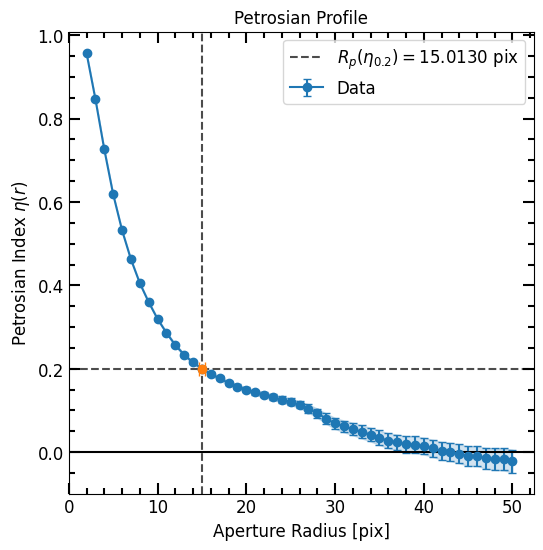
In the Photometry Chapter we constructed a curve of growth for the football shaped galaxy:
[15]:
import numpy as np
from astropy.table import Table
phot_table = Table.read('data/abell_2744_galaxy_f105w_photometry.ecsv') # Read table
# Load data
r_list = np.array(phot_table['r_list'])
flux_arr = np.array(phot_table['flux_arr'])
area_arr = np.array(phot_table['area_arr'])
error_arr = np.array(phot_table['error_arr'])
# Make Petrosian profile
p = pf.Petrosian(r_list, area_arr, flux_arr, flux_err=error_arr)
p.plot()
plt.show()
Correct Profile:
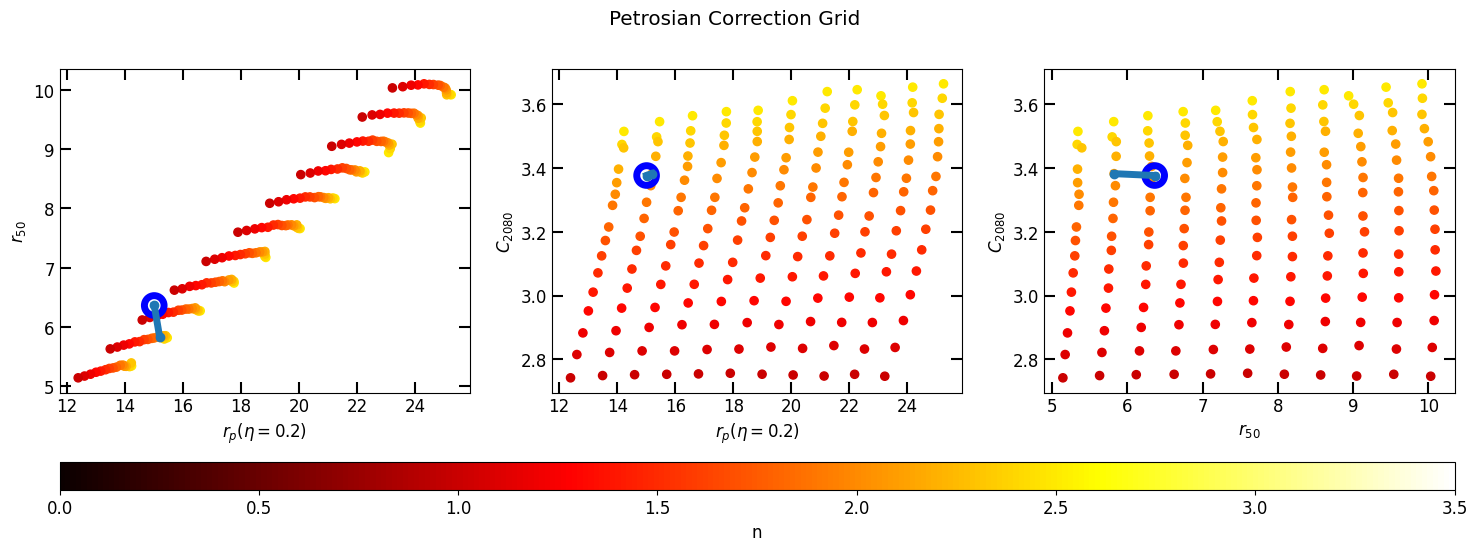
Simply use the
correct(p)function to correct the profile.Use
plot_correctionfunction to see the closest value.The parameters used for correction are derived directly from the grid. Specifically:
'p02': Represents the Petrosian radius \(P_{02} = R(\eta_{0.2})\).'u_r_50': Stands for the uncorrected half-light radius.'u_c2080': Corresponds to the uncorrected concentration index.
[16]:
pc = pf.PetrosianCorrection.read('data/f105w_psf_corr.ecsv')
# Plot grid and uncorrected profile on grid (p)
pc.plot_correction(p)
plt.show()
# Pass uncorrected p to the correct function
p_corrected = pc.correct(p)
# Plot corrected profile
p_corrected.plot()
plt.show()
Estimating Parameters:
The method
estimate_n(p)will give an estimated Sérsic indexnbased on the half-light radius andc2080computed using the default epsilon value.The method
estimate_epsilon(p)will provide a corrected epsilon value given the half-light radius andc2080computed with the default epsilon.
[17]:
n = pc.estimate_n(p)
n
[17]:
2.1
[18]:
corr_epsilon = pc.estimate_epsilon(p)
corr_epsilon
[18]:
2.227805629021434