Multi-object Photometry
In this section we demonstrate how to perform photometry on multiple objects in an image. We choose a faint group of galaxies and run the photometry steps as described in the Photometry section.
To start with PetroFit, simply import it as follows:
[1]:
import petrofit as pf
Loading Example Data
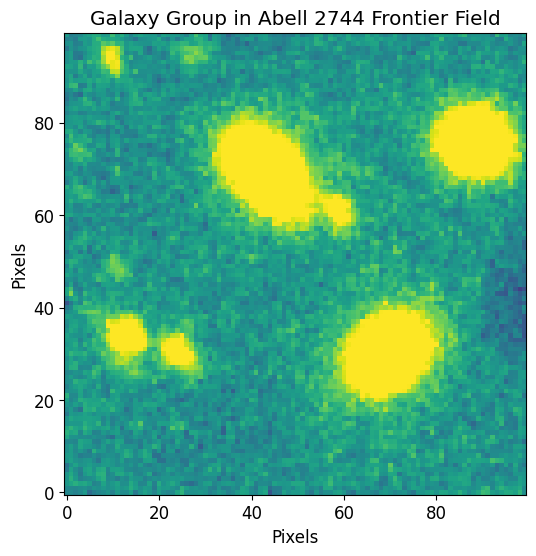
The following data is a cutout of a group of faint galaxies in Abell 2744. The original data was acquired by the Hubble Frontier Fields team via the WFC3 instrument in the F105W filter and can be directly downloaded from the Mikulski Archive for Space Telescopes.
We first use astropy’s CCDData to load the example data and visualize it through matplotlib.
[2]:
from astropy.nddata import CCDData
image = CCDData.read('data/abell_2744_group_of_galaxies_f105w.fits.gz')
[4]:
from matplotlib import pyplot as plt
plt.rcParams['figure.figsize'] = [6, 6]
plt.rcParams['image.origin'] = 'lower'
plt.rcParams['font.size'] = 12
vmax = 0.01 # Use the image std as max and min of all plots
vmin = - vmax
plt.imshow(image.data, vmin=vmin, vmax=vmax)
plt.title("Galaxy Group in Abell 2744 Frontier Field")
plt.xlabel("Pixels")
plt.ylabel("Pixels")
plt.show()
Make Source Catalog
We will use the sigma clipped std as a threshold at the segmentation and deblending steps.
[5]:
from astropy.stats import sigma_clipped_stats
# Compute stats for threshold
image_mean, image_median, image_stddev = sigma_clipped_stats(image.data, sigma=3)
# Define threshold
threshold = image_stddev
# Min Source size (area)
npixels = 5**2
cat, segm, segm_deblend = pf.make_catalog(
image.data,
threshold=threshold,
wcs=image.wcs,
deblend=True,
npixels=npixels,
contrast=0.00,
plot=True, vmax=vmax, vmin=vmin,
figsize=[12, 6]
)
plt.show()
# Display source properties
print("Num of Targets:", len(cat))
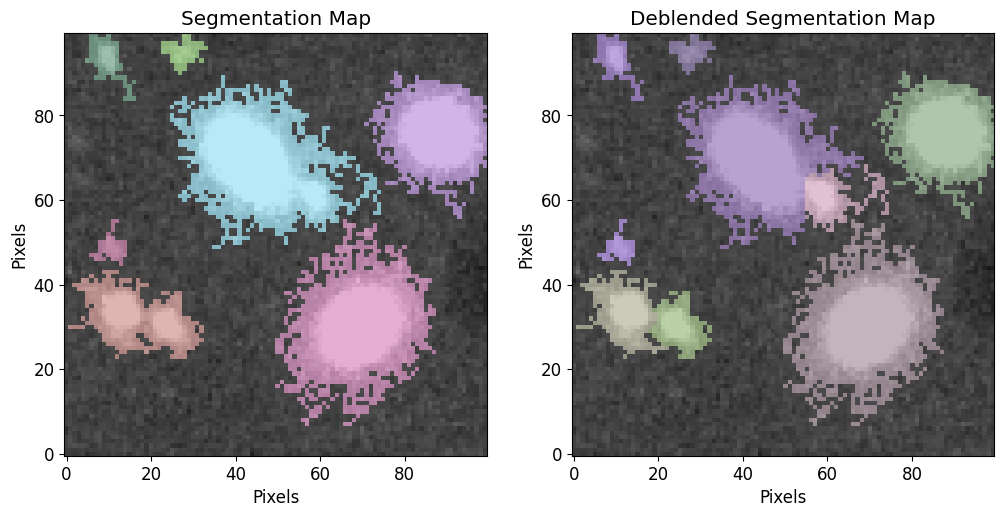
Num of Targets: 9
Photometry Loop
We define the list of aperture radii and proceed to the photometry step. In this case, instead of selecting a source, we loop through the source catalog and perform photometry on each object. After constructing the photometry we create a Petrosian object for the source. We save the Petrosian in a python dictionary (petrosian_properties) for later use.
[6]:
max_pix=35
r_list = pf.make_radius_list(
max_pix=max_pix, # Max pixel to go up to
n=max_pix # the number of radii to produce
)
petrosian_properties = {}
for idx, source in enumerate(cat):
# Photomerty
flux_arr, area_arr, error_arr = pf.source_photometry(
# Inputs
source, # Source (`photutils.segmentation.catalog.SourceCatalog`)
image.data, # Image as 2D array
segm_deblend, # Deblended segmentation map of image
r_list, # list of aperture radii
# Options
cutout_size=max(r_list)*2, # Cutout out size, set to double the max radius
bg_sub=True, # Subtract background
sigma=3, sigma_type='clip', # Fit a 2D plane to pixels within 3 sigma of the mean
plot=False, vmax=vmax, vmin=vmin, # Show plot with max and min defined above
)
plt.show()
p = pf.Petrosian(r_list, area_arr, flux_arr)
petrosian_properties[source] = p
print("Completed for {} Sources".format(len(petrosian_properties)))
Completed for 9 Sources
Compute Total Mags
We compute the total magnitudes of the sources by looping through their Petrosian objects. We use the flux_to_abmag function to convert the total flux to AB mags of each object.
[7]:
import numpy as np
mag_list = []
for source in petrosian_properties:
# Get Petrosian
p = petrosian_properties[source]
# Compute HST Flux -> mags for total_flux
mag = pf.hst_flux_to_abmag(p.total_flux, image.header)
# Add to mag list
mag_list.append(mag)
# Convert mag_list to array
mag_list = np.array(mag_list)
mag_list
[7]:
array([23.78101354, 28.64431073, 24.01132477, 27.41390818, 28.64179264,
26.41465787, 25.84712754, 23.55719409, 26.37657836])
Photometry Catalog
We construct and save a photometry catalog with the magnitudes we computed. To construct the table, we first use the SourceCatalog.to_table() function that returns an astropy table. This will include important info about each source. We then add a new column (MAG_F105W) with the total mags we computed.
[8]:
# Segmentation catalog to astropy table.
photo_cat = cat.to_table()
# Add new column with mags.
photo_cat.add_column(mag_list, index=4, name='MAG_F105W')
# Save to file.
photo_cat.write('temp/example_photo_cat.csv', overwrite=True)
photo_cat
[8]:
| label | xcentroid | ycentroid | sky_centroid | MAG_F105W | bbox_xmin | bbox_xmax | bbox_ymin | bbox_ymax | area | semimajor_sigma | semiminor_sigma | orientation | eccentricity | min_value | max_value | local_background | segment_flux | segment_fluxerr | kron_flux | kron_fluxerr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg,deg | pix2 | pix | pix | deg | ||||||||||||||||
| int64 | float64 | float64 | SkyCoord | float64 | int64 | int64 | int64 | int64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 |
| 1 | 69.32265836375497 | 30.238019971715627 | 3.579223902860569,-30.380187463851975 | 23.7810135425286 | 50 | 87 | 7 | 55 | 1000.0 | 7.066388328760255 | 5.579720371665397 | 54.75734563367578 | 0.6136036659242836 | 0.0018545736093074083 | 0.07670669257640839 | 0.0 | 10.244122936739586 | nan | 10.709215362358297 | nan |
| 2 | 10.737608995340572 | 48.6121151841155 | 3.580355742519194,-30.379881269181666 | 28.644310734022653 | 6 | 14 | 45 | 55 | 43.0 | 2.0965997789103628 | 1.8762638664800115 | 73.19191801040047 | 0.4462507287495157 | 0.0019008400849997997 | 0.00620837789028883 | 0.0 | 0.15271575504448265 | nan | 0.395127586728858 | nan |
| 3 | 87.92829837715828 | 75.63296420231474 | 3.5788644863585364,-30.37943086630141 | 24.011324773909678 | 71 | 99 | 56 | 90 | 569.0 | 4.684225490343459 | 4.371347211905419 | -44.65368081791386 | 0.3593419120981481 | 0.001854302710853517 | 0.10125063359737396 | 0.0 | 8.084702808177099 | nan | 8.19241690061656 | nan |
| 4 | 9.815810702861459 | 93.31562288938129 | 3.580373573168226,-30.37913621099525 | 27.413908182788763 | 5 | 16 | 84 | 99 | 77.0 | 3.355176286646171 | 1.6479017677110668 | -61.23856024451831 | 0.8710738471935022 | 0.0018753919284790754 | 0.012467581778764725 | 0.0 | 0.3568276413716376 | nan | 0.46358781728153653 | nan |
| 5 | 27.555493767397174 | 94.91373210653953 | 3.5800308569397217,-30.37910956468503 | 28.641792635176103 | 23 | 33 | 90 | 99 | 54.0 | 2.252178183265229 | 1.8456062428385482 | 9.760103655188653 | 0.5731133274324416 | 0.0018675385508686304 | 0.006537159439176321 | 0.0 | 0.18736975535284728 | nan | 0.21081410418602697 | nan |
| 6 | 24.481221829558184 | 30.35265489108243 | 3.5800902145053146,-30.38018558504191 | 26.414657874624123 | 18 | 32 | 23 | 38 | 137.0 | 3.1297440100684626 | 2.368327034962023 | -54.918957106073215 | 0.6537438768702845 | 0.0018714345060288906 | 0.02676144801080227 | 0.0 | 0.9589369829045609 | nan | 1.161363705896545 | nan |
| 7 | 12.418600181421457 | 33.41806848906761 | 3.5803232591739147,-30.380134502367362 | 25.84712753793599 | 1 | 22 | 23 | 43 | 221.0 | 4.158127754208604 | 3.041444861227075 | -54.91674752060506 | 0.6818996401383097 | 0.0018537540454417467 | 0.02450975775718689 | 0.0 | 1.475870828377083 | nan | 1.9072046957775326 | nan |
| 8 | 43.420003300978024 | 69.4761061324968 | 3.57972435145291,-30.379533514551756 | 23.557194090925286 | 25 | 68 | 49 | 89 | 878.0 | 6.259160655628094 | 4.972810345866363 | -47.74028219223737 | 0.607283547825944 | 0.0018594693392515182 | 0.13125701248645782 | 0.0 | 12.524548709043302 | nan | 12.935097062743418 | nan |
| 9 | 59.890633529332646 | 60.66274815897566 | 3.5794061443723244,-30.379680392014183 | 26.376578364563656 | 55 | 74 | 53 | 68 | 144.0 | 3.985926639607556 | 2.811090552294893 | 0.7501050272960306 | 0.7089547587670784 | 0.001866802922450006 | 0.02579653263092041 | 0.0 | 0.9505528409499675 | nan | 1.816363676057363 | nan |
Simultaneous Fitting
In this section, we explore how to make compound models that can be used to describe multiple objects in an image. We use the same dataset as the Multi-object Photometry section to fit the nine faint sources with Sersic profiles.
Make Individual Models
To do this we loop through the sources and construct astropy Sersic2D models for each source. We also make initial guesses of the paramters as described in the Estimating Sersic Parameters section. At the end of each iteration, we add the newly constructed model in a list model_list.
[9]:
from photutils.isophote import EllipseGeometry, Ellipse
from astropy.modeling import models
# AstroPy Model List
model_list = []
# For each source
for source in list(petrosian_properties.keys()):
# Get Petrosian
p = petrosian_properties[source]
# Estimate center
position = pf.get_source_position(source)
x_0, y_0 = position
# Estimate shape
elong = pf.get_source_elong(source)
ellip = pf.get_source_ellip(source)
theta = pf.get_source_theta(source)
# Estimate Sersic index
n = 1
# Estimate r_half_light
r_eff = p.r_half_light
# Estimate amplitude
amplitude = pf.get_amplitude_at_r(r_eff, image, x_0, y_0, ellip, theta)
# Allow for 4 pixel center slack
center_slack = 4
# Make astropy model
sersic_model = models.Sersic2D(
amplitude=amplitude,
r_eff=r_eff,
n=n,
x_0=x_0,
y_0=y_0,
ellip=ellip,
theta=theta,
bounds = pf.get_default_sersic_bounds({
'x_0': (x_0 - center_slack/2, x_0 + center_slack/2),
'y_0': (y_0 - center_slack/2, y_0 + center_slack/2),
}),
)
# Add to model list
model_list.append(sersic_model)
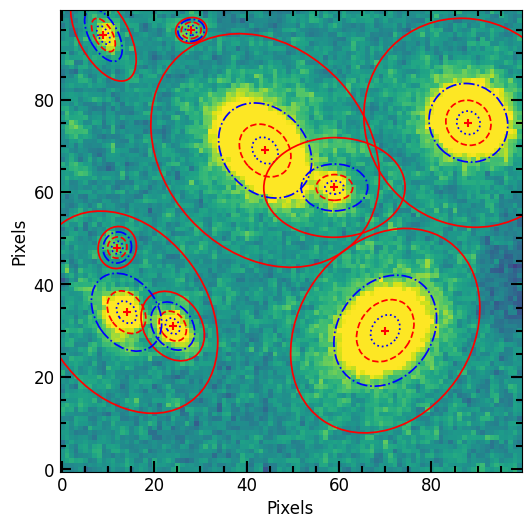
# Over-plot Petrosian radii
p.imshow(position=position, elong=elong, theta=theta, lw=1.25)
# Plot image of sources
plt.imshow(image.data, vmax=vmax, vmin=vmin)
pf.mpl_tick_frame()
plt.xlabel('Pixels')
plt.ylabel('Pixels')
plt.show()
Make Compound Model
To make a single compound model that represents all the sources of interest, we add up all the models. astropy models can be added like numbers or arrays, so we convert the model list to a numpy array and sum it.
[10]:
compound_model = np.array(model_list).sum()
compound_model
[10]:
<CompoundModel(amplitude_0=0.02231892, r_eff_0=7.18243649, n_0=1., x_0_0=70., y_0_0=30., ellip_0=0.21038583, theta_0=0.95569597, amplitude_1=0.00356518, r_eff_1=2.31946389, n_1=1., x_0_1=12., y_0_1=48., ellip_1=0.10509202, theta_1=1.27743996, amplitude_2=0.03006719, r_eff_2=5.04680936, n_2=1., x_0_2=88., y_0_2=75., ellip_2=0.06679403, theta_2=-0.77935375, amplitude_3=0.00509282, r_eff_3=3.97219444, n_3=1., x_0_3=9., y_0_3=94., ellip_3=0.50884793, theta_3=-1.06881451, amplitude_4=0.0044909, r_eff_4=2.15623125, n_4=1., x_0_4=28., y_0_4=95., ellip_4=0.18052388, theta_4=0.17034594, amplitude_5=0.01000131, r_eff_5=3.48929786, n_5=1., x_0_5=24., y_0_5=31., ellip_5=0.24328411, theta_5=-0.95851662, amplitude_6=0.00696394, r_eff_6=5.01960392, n_6=1., x_0_6=14., y_0_6=34., ellip_6=0.26855425, theta_6=-0.95847806, amplitude_7=0.03945622, r_eff_7=6.24384877, n_7=1., x_0_7=44., y_0_7=69., ellip_7=0.20551483, theta_7=-0.83322511, amplitude_8=0.00847777, r_eff_8=3.95179036, n_8=1., x_0_8=59., y_0_8=61., ellip_8=0.29474604, theta_8=0.0130918)>
Make PSFConvolvedModel2D
Now that we have a single model that represents all the sources, we can create a PSFConvolvedModel2D with the appropriate parameters. After we load the PSF, we wrap the compound model and PSF using PSFConvolvedModel2D. We specify an oversampling factor 4 to account for poor CCD sampling.
Load and Normalize PSF
[11]:
from astropy.io import fits
PSF = fits.getdata('data/f105w_psf.fits.gz')
PSF = PSF / PSF.sum()
plt.imshow(PSF, vmin=0, vmax=PSF.std()/10)
[11]:
<matplotlib.image.AxesImage at 0x7f8035703f90>
PSFConvolvedModel2D
[12]:
psf_sersic_model = pf.PSFConvolvedModel2D(compound_model, psf=PSF, oversample=4)
psf_sersic_model.fixed['psf_pa'] = True
Fit Model to Data
We fit the compound model using a Levenberg-Marquardt algorithm and save the returned optimized copy of the fitted model in fitted_model. Since this the compound model is composed of many parameters, we may see astropy warnings when the fitter explores parameters that cause issues, such as division by zero.
[13]:
%%time
fitted_model, _ = pf.fit_model(
image.data, psf_sersic_model,
maxiter=10000,
epsilon=1.4901161193847656e-08,
acc=1e-09,
)
CPU times: user 2min 26s, sys: 7.98 s, total: 2min 34s
Wall time: 2min 22s
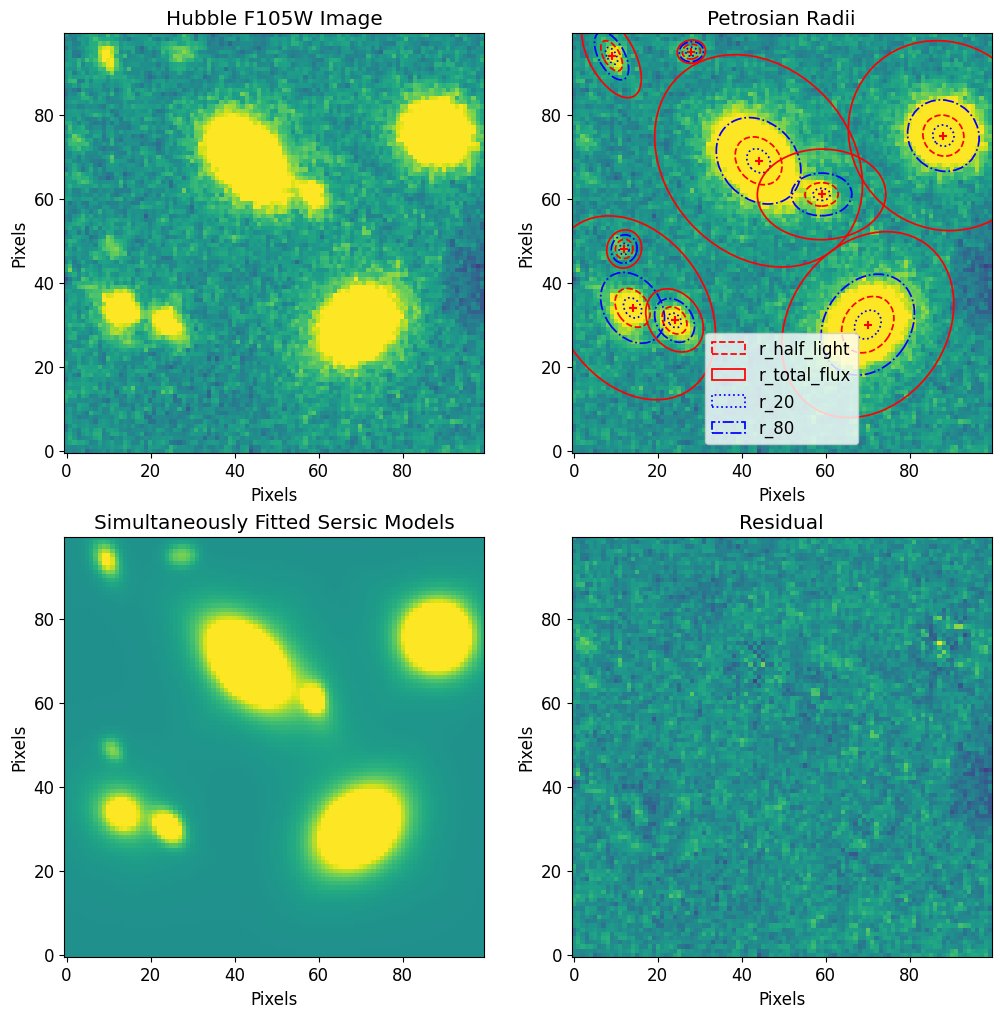
Generate Model Image
To generate a model image we use the model_to_image utility function. This function allows us to define the center of the model image and the side length of the image.
[14]:
# Make Model Image
# ----------------
# Set the size of the model image equal to the original image
full_fitted_image_size = image.data.shape[0]
# Center the model image at the center of the original image
# so the two images cover the same window
full_fitted_image_center = full_fitted_image_size // 2
# Generate a model image from the model
fitted_model_image = pf.model_to_image(
fitted_model,
full_fitted_image_size,
)
# Plot Model Image
# ----------------
fig, ax = plt.subplots(2, 2, figsize=[12, 12])
# Plot raw data
im = ax[0, 0].imshow(image.data, vmin=vmin, vmax=vmax)
ax[0, 0].set_title("Hubble F105W Image")
ax[0, 0].set_xlabel("Pixels")
ax[0, 0].set_ylabel("Pixels")
#ax[0, 0].axis('off')
# Plot Petrosian radii
plt.sca(ax[0, 1])
for i, source in enumerate(petrosian_properties):
p = petrosian_properties[source]
position = pf.get_source_position(source)
x_0, y_0 = position
elong = pf.get_source_elong(source)
ellip = pf.get_source_ellip(source)
theta = pf.get_source_theta(source)
p.imshow(position=position, elong=elong, theta=theta, lw=1.25)
if i == 0:
plt.legend()
ax[0, 1].imshow(image.data, vmin=vmin, vmax=vmax)
ax[0, 1].set_title("Petrosian Radii")
ax[0, 1].set_xlabel("Pixels")
ax[0, 1].set_ylabel("Pixels")
# ax[0, 1].axis('off')
# Plot Model Image
ax[1, 0].imshow(fitted_model_image, vmin=vmin, vmax=vmax)
ax[1, 0].set_title("Simultaneously Fitted Sersic Models")
ax[1, 0].set_xlabel("Pixels")
ax[1, 0].set_ylabel("Pixels")
# ax[1, 0].axis('off')
# Plot Residual
ax[1, 1].imshow(image.data - fitted_model_image, vmin=vmin, vmax=vmax)
ax[1, 1].set_title("Residual")
ax[1, 1].set_xlabel("Pixels")
ax[1, 1].set_ylabel("Pixels")
# ax[1, 1].axis('off')
plt.show()
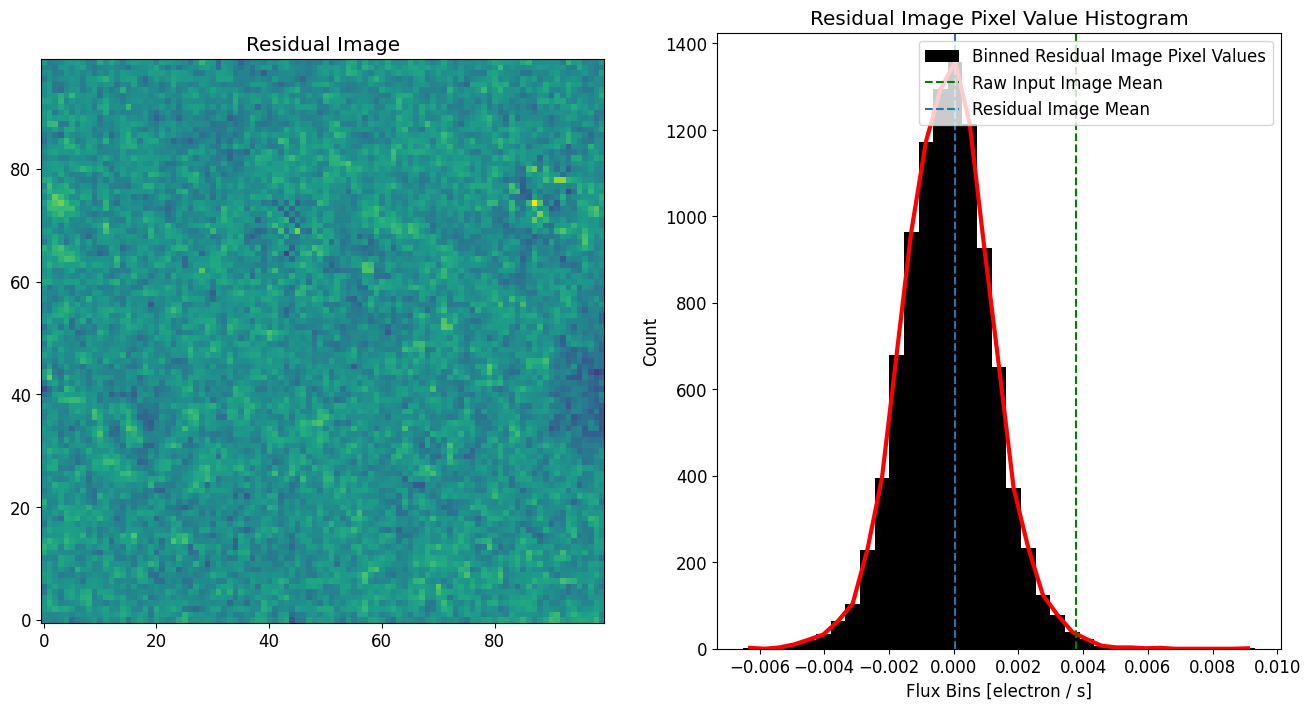
Analysis of Background
We can now create a background image using the residual and perform some statistical analysis on it.
[15]:
# Define background image
background_image = image.data - fitted_model_image
# Compute stats
# -------------
noise_mean = background_image.mean()
noise_sigma = background_image.std()
noise_3_sigma = noise_sigma * 3.
noise_8_sigma = noise_sigma * 8.
print("noise_mean = {}".format(noise_mean))
print("noise_sigma = {}".format(noise_sigma))
print("noise_3_sigma = {}".format(noise_3_sigma))
# Plots
# -----
fig, axs = plt.subplots(1, 2, figsize=[16, 8])
plt.sca(axs[0])
plt.imshow(image.data - fitted_model_image, vmin=vmin, vmax=vmax)
plt.title('Residual Image')
plt.sca(axs[1])
n, bins, patches = plt.hist(background_image.flatten(), bins=35, align='left',
color='black', label="Binned Residual Image Pixel Values")
plt.plot(bins[:-1], n, c='r', linewidth=3)
plt.axvline(image.data.mean(), label="Raw Input Image Mean", c='g',linestyle="--")
plt.axvline(noise_mean, label="Residual Image Mean", linestyle="--")
plt.xlabel('Flux Bins [{}]'.format(str(image.unit)))
plt.ylabel('Count')
plt.title('Residual Image Pixel Value Histogram')
plt.legend()
plt.show()
noise_mean = 5.009439712276197e-05
noise_sigma = 0.0013882674260103844
noise_3_sigma = 0.004164802278031153
Now we see the residual image mean is near that is near the mean of the noise distribution, we can make a segmentation map using the residual image 3-sigma as the detection threshold. Notice how some of the sources we were able to fit were below the 3-sigma estimate of the background (residual image).
[16]:
new_threshold = noise_3_sigma
new_cat, new_segm, new_segm_deblend = pf.make_catalog(
image.data,
threshold=new_threshold,
wcs=image.wcs,
deblend=True,
npixels=npixels,
contrast=0.00,
plot=True, vmax=vmax, vmin=vmin,
figsize=[12, 6]
)
plt.show()
# Display source properties
print("Num of Targets:", len(new_cat))
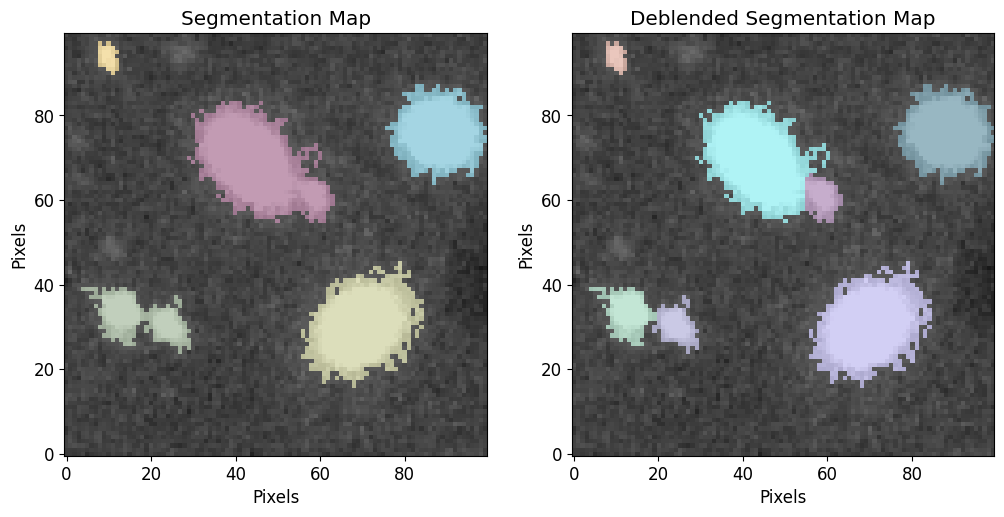
Num of Targets: 7